RNAimmuno Help
1. Who are we?2. About
3 .Searching the RNAimmuno database
3.1 Browse
3.2 Quick search
3.3 Advanced search
Reagent name
Reagent sequence
4. Description of activation/induction status
5. RNAimmuno tools
6. Abbreviations
7. Implementation
1. Who are we?
RNAimmuno is managed by researchers at the Department of Molecular Biomedicine, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznan, Poland
RNAimmuno Team:
Questions or comments?
E-mail rnaimm@ibch.poznan.pl
RNAimmuno is managed by researchers at the Department of Molecular Biomedicine, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznan, Poland
RNAimmuno Team:
- Marta Olejniczak
- Paulina Galka-Marciniak
- Katarzyna Polak
- Martyna Urbanek
- Grzegorz Jankowiak
- Wlodzimierz J. Krzyzosiak
Questions or comments?
E-mail rnaimm@ibch.poznan.pl
2. About
RNAimmuno is manually curated database which has been developed to gather information concerning non-specific immunological effects of RNAi triggers and miRNA regulators. The database collects the published data describing immunological side effects in cells and model organisms caused by different types of reagents, eg. siRNAs, shRNAs, sh-miRs and antimiRs. Also, it contains the most important information about known cellular sensors of foreign RNA and DNA (SENSORS button) and the pathways these sensors activate (PATHWAYS button). All the information is presented in tabular format with links to external public databases.
The existing data indicate that not only the reagents but also their carriers induce immune responses. Considering this, we have included published data about the toxicity of transfection reagents frequently used in RNAi experiments (DELIVERY/UPTAKE button).
Exemplary applications of the RNAimmuno database are shown below:
RNAimmuno is manually curated database which has been developed to gather information concerning non-specific immunological effects of RNAi triggers and miRNA regulators. The database collects the published data describing immunological side effects in cells and model organisms caused by different types of reagents, eg. siRNAs, shRNAs, sh-miRs and antimiRs. Also, it contains the most important information about known cellular sensors of foreign RNA and DNA (SENSORS button) and the pathways these sensors activate (PATHWAYS button). All the information is presented in tabular format with links to external public databases.
The existing data indicate that not only the reagents but also their carriers induce immune responses. Considering this, we have included published data about the toxicity of transfection reagents frequently used in RNAi experiments (DELIVERY/UPTAKE button).
Exemplary applications of the RNAimmuno database are shown below:
- User may check whether a reagent of interest, or similar reagents, delivered with the use of a selected carrier activated the immune response in a specific experimental model.
- User may also be able to find a less toxic reagent and the most suitable experimental conditions (e.g., reagent concentration and delivery method) to silence the selected target gene.
- The RNAimmuno database provides an opportunity to find a proper control reagent which has been characterized with regard to the induction of non-sequence-specific effects.
- Through the use of specific filtering options, it is possible to analyze the influence of various parameters (e.g., reagent type, sequence, concentration, and chemical modifications) on the activation of specific cellular sensors and on cell viability.
- To identify RNA reagents that strongly activate the immune system which may be especially important in e.g., antiviral and cancer therapy studies.
- "Simple tool" option allows scanning the reagent of interest for the presence of the most important molecular patterns known to activate the cellular sensors of foreign RNA (immunostimulatory motifs, reagent’s length, blunt end and 5’triphosphate).
- "A more advanced “sequence alignment" option may be used to asses degree of homology between query sequence and reagents described in the database.
3. Searching the RNAimmuno database<
You may either use the quick search window to enter a keyword or choose specific records from the drop-down list of records in the advanced search.
Browse
All records can be displayed by using the “browse” option from the top panel in the home page.
Quick search
Enter a keyword:
Advanced search
User specifies:
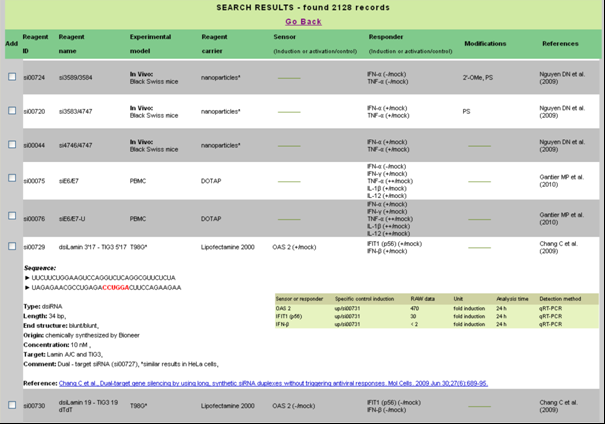
As a result of such search users are given a tabular set of information with links to publications from which the data were extracted. The main table of results contains data concerning reagent ID and original name, experimental model, reagent carrier, the level of induction/activation of foreign RNA sensors and responders, and the chemical modification of the reagent when specified. Particular records contain drop-down detailed information which include reagent type, its sequence, length, end structure, origin, concentration, target gene, observed effects (raw data) and comment. Data which need additional comment are marked with asterisks. User may also choose specific records and display them in a new window (see Figure below).
Reagent name
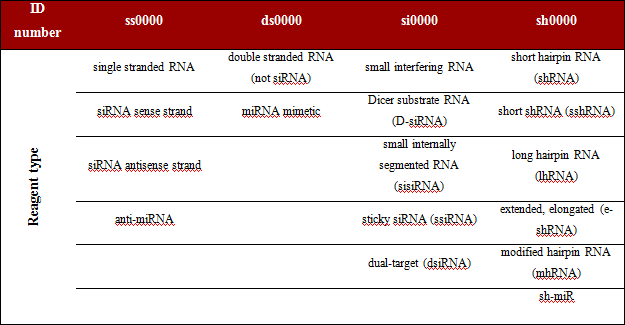
In the current version of the database all reagents contain their names given in the original publications along with their type and specific ID number that makes each reagent easy distinguishable from others. All types of reagents are divided into 4 groups (columns in Table 1 below) and an abbreviated name of each group (ss, ds, si and sh) precedes the ID number, e.g. ss00001; ds00002; si00003; sh00004. Generally the very last ID number in our database indicates the total number of sequences included. The same reagents analyzed in different experimental conditions (e.g. cell types, delivery methods) may cause different effects, therefore, they are distinguished as different records (with the same reagent ID). On the other hand, when a given reagent was used at different concentrations it has been described as a single record (results were given as a range of raw values). RNAimmuno v.1.0 database contains over 2,100 records based on over 1,300 unique sequences.
Table 1. Groups of reagents and their prefixes in ID numbers.
Reagent sequence
You may either use the quick search window to enter a keyword or choose specific records from the drop-down list of records in the advanced search.
Browse
All records can be displayed by using the “browse” option from the top panel in the home page.
Quick search
Enter a keyword:
- protein symbol (e.g. PKR, TLR3, IFN)
- reagent type (e.g. siRNA, shRNA)
- reagent carrier (e.g. Lipofectamine, DOTAP, Lentivirus)
- experimental model (e.g. HEK, HeLa, mice)
- reagent ID number
- target gene
- name of the first author of relevant publication
Advanced search
User specifies:
- reagent type
- reagent carrier
- experimental model
- sensor (protein that recognizes foreign RNA or DNA)
- responder (i.e. proteins from the activated pathway e.g., transcription factors, cytokines, or chemokines which function downstream of cellular sensors recognizing foreign RNA or DNA)
- reagent modifications (chemical modifications) – modified nucleotides are shown in parenthesis
- known immunostimulatory motifs
- level of activation/induction
- reagent sequence (5’→ 3’) – selected sequence is highlighted in red
- name of the first author of relevant publication
As a result of such search users are given a tabular set of information with links to publications from which the data were extracted. The main table of results contains data concerning reagent ID and original name, experimental model, reagent carrier, the level of induction/activation of foreign RNA sensors and responders, and the chemical modification of the reagent when specified. Particular records contain drop-down detailed information which include reagent type, its sequence, length, end structure, origin, concentration, target gene, observed effects (raw data) and comment. Data which need additional comment are marked with asterisks. User may also choose specific records and display them in a new window (see Figure below).

Reagent name
In the current version of the database all reagents contain their names given in the original publications along with their type and specific ID number that makes each reagent easy distinguishable from others. All types of reagents are divided into 4 groups (columns in Table 1 below) and an abbreviated name of each group (ss, ds, si and sh) precedes the ID number, e.g. ss00001; ds00002; si00003; sh00004. Generally the very last ID number in our database indicates the total number of sequences included. The same reagents analyzed in different experimental conditions (e.g. cell types, delivery methods) may cause different effects, therefore, they are distinguished as different records (with the same reagent ID). On the other hand, when a given reagent was used at different concentrations it has been described as a single record (results were given as a range of raw values). RNAimmuno v.1.0 database contains over 2,100 records based on over 1,300 unique sequences.
Table 1. Groups of reagents and their prefixes in ID numbers.

Reagent sequence
- Reagent sequences are displayed in 5’→3’ direction.
- Immunostimulatory motifs selected from the list or sequences specified by user are highlighted in red.
- Chemically modified nucleotides are shown in parenthesis.
- Nucleotides that form loop structure within hairpin – types reagents (e.g. shRNAs, sh-miRs) are lower cased.
4. Description of activation/induction status
We found it difficult to apply a universal scale of activation effects due to the use of different experimental conditions, methods of analysis and biological systems by different authors (e.g. in vivo systems vs in cell lines). Therefore, the scale we used to describe the observed effects: no activation/induction (-/mock), activation/induction (+/mock) and strong activation/induction (++/mock) generally refers to the results described in specific publications (obtained in specific experimental conditions).
Additionally, the activation status of the sensor or responder is presented as an activation/induction relevant to specific control (up/specific control, down/specific control, similar/specific control), and wherever possible also the fold-change is indicated (e.g. 5 fold up/specific control). To avoid any misinterpretation of the results the raw data from the original publications are also included.
5. RNAimmuno tools
The database provides the user with tools that may be helpful in designing safer RNAi reagents. The "simple tool" option allows scanning the query sequence for the presence of the most important molecular patterns known to activate the cellular sensors of foreign RNA (immunostimulatory motifs, reagent's length, blunt end and 5'triphosphate). A more advanced tool uses BLASTN algorithm (release 2.2.25) to align input sequence with reagents collected in the RNAimmuno database. Currently this tool operates on default parameters that are optimal for short input sequences, but in the future we plan to develop a more advanced option by changes in BLASTN parameters or by using SSEARCH. The results of a search are summarized in a simple tabular format of hits, followed by alignments of the query sequence against each hit sequence. The parameters of the search are summarized at the bottom of the page. The minimal length of the query sequence is 10 nucleotides. Detailed information about the effects generated by reagents showing a high degree of similarity to the query sequence is accessible via specific links in the table.
6. Abbreviations
Reagent Type
| anti-miR | antago-miR, microRNA antagonist |
| dsiRNA | dual-target siRNA |
| D-siRNA | Dicer substrate siRNA |
| dsRNA | double-stranded RNA, the records in this category of search correspond to reagents that were defined by authors as dsRNA, however, in some cases they are typical siRNAs |
| e-shRNA | extended short hairpin RNA |
| lhRNA | long hairpin RNA |
| mhRNA | modified hairpin RNA |
| miRNA mimetic | synthetic miRNAv |
| sh-miR | pri-miRNA-based short hairpin RNA |
| shRNA | short hairpin RNA |
| siRNA | small interfering RNA |
| sisiRNA | small internally segmented RNA |
| sshRNA | short shRNA (hairpins with a stem length of 19bp or less) |
| ssiRNA | sticky overhang siRNA |
| ssRNA | single-stranded RNA, the records in this category of search correspond to reagents that were defined by authors as ssRNAs, |
| ss siRNA | single-stranded siRNA (one strand of the siRNA duplex) |
End structure
| 3’NN/3’NN | dinucleotide overhangs at both 3’ends |
| blunt/3’NN | dinucleotide overhang only at one 3’end |
| blunt/blunt | duplex without overhangs |
| 3’ 8N/3’ 5N | the number of Ns indicates the length of overhang |
| 3’Un | undefined length of the uridine tail in the shRNA |
| 5’ppp | triphosphate group at the 5’ end |
| 5’ppp, 3’NN/3’NN | triphosphate group at the 5’ end, dinucleotide overhangs at both 3’ends |
Modifications - modified nucleotides are shown in parenthesis
| 2’F | 2'-fluoro |
| 2'-OMe | 2’-O-methyl |
| ADA | 2'-N-adamant-1-ylmethylcarbonyl-2'-amino-LNA |
| AEM | 2'-aminoethoxymethyl |
| AENA | 2'-deoxy-2'-N,4'-C-ethylene-LNA |
| ALN | alfa-L-LNA |
| ANA | altritol nucleic acid |
| AP | 2'-aminopropyl |
| APM | 2'-aminopropoxymethyl |
| CE | 2'-cyanoethyl |
| CENA | 2',4'-carbocyclic-ENA-LNA |
| CLNA | 2',4'-carbocyclic-LNA-locked nucleic acid |
| EA | 2'-aminoethyl |
| FITC | fluorescein isothiocyanate |
| GE | 2'-guanidinoethyl |
| HM | 4'-C-hydroxymethyl-DNA |
| HNA | hexitol nucleic acid |
| HS-backbone | |
| I | inosine |
| LNA | locked nucleic acid |
| OX | oxetane-LNA |
| PS | phosphorothioate linkage |
| PYR | 2'-N-pyren-1-ylmethyl-2'-amino-LNA |
| UNA | unlocked nucleic acid |
Delivery
| HPTV | tail vein injection via high-pressure |
| i.v. | intravenous |
| IP | intraperitoneally |
| LPTV | tail vein injection via low-pressure |
| PEI | polyethyleneimine |
| pLa | poly-L-arginine |
| PLL | poly-L-lysine |
| PTD-DRBD | Peptide Transduction-dsRNA Binding Domain |
| SNALP | Stable Nucleic Acid-Lipid Particles |
7. Implementation
The RNAimmuno database (v. 1.0) runs in a Linux environment, and it has been developed as a relational database in MySQL. The search engine is served by the Apache http daemon along with the PHP scripts. The interface component consists of the web pages designed and implemented in HTML/CSS. It has been tested in the latest version of web browsers, including Mozilla Firefox, Internet Explorer, Opera, Safari, and Google Chrome. The service is hosted and maintained by the Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznan, Poland.